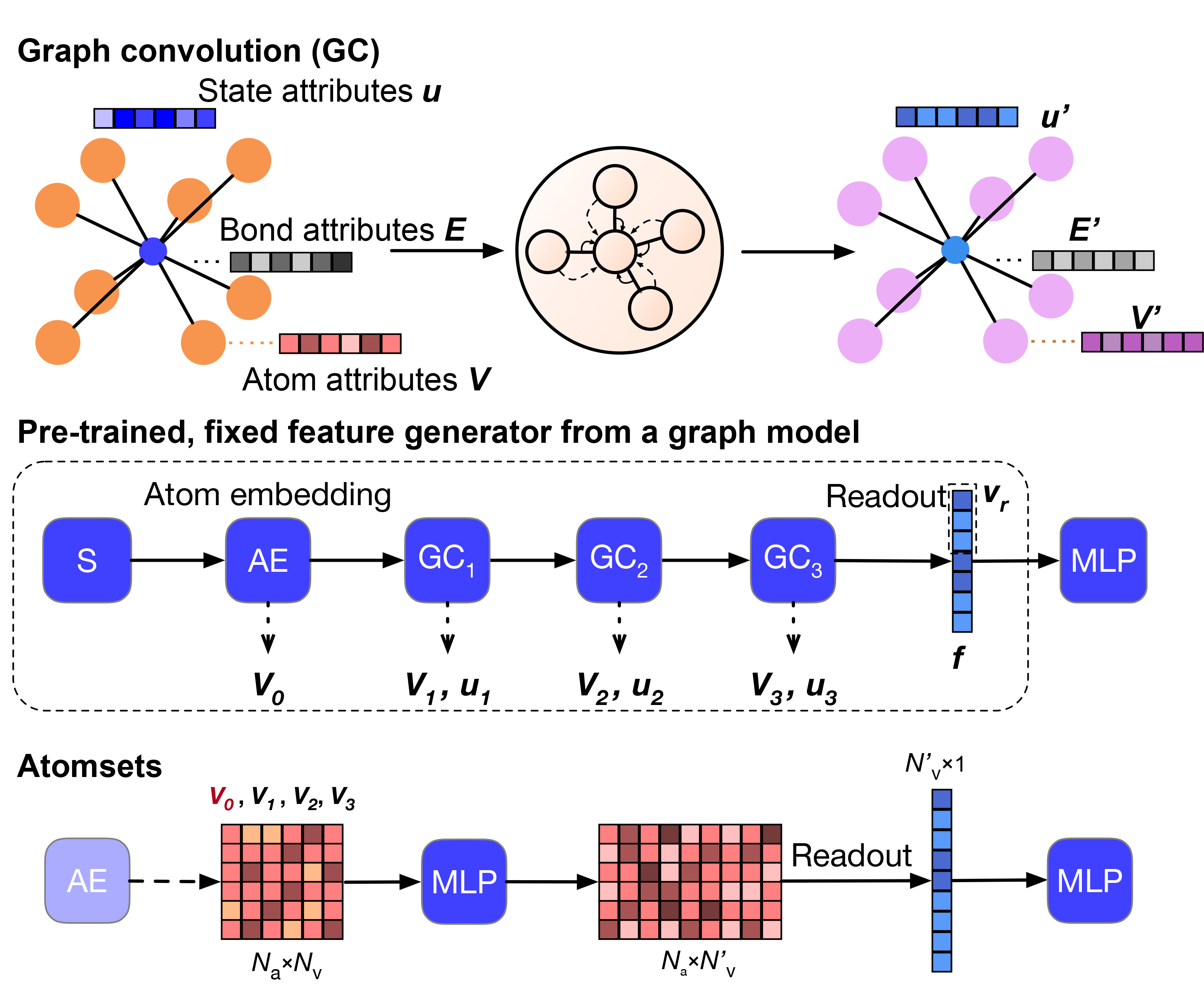
AtomSets – using graph networks as an encoder
Graph networks are an extremely powerful deep learning tool for predicting materials properties. However, a critical weakness is their reliance on large quantities of training data. In this work published in npj Computational Materials, Dr Chi Chen shows that pre-trained MEGNet formation energy models can be effectively used as “encoders” for crystals in what we call the AtomSets framework. The compositional and structural descriptors extracted from graph network deep learning models, combined with standard artificial neural network models, can achieve lower errors than the graph network models at small data limits and other non-deep-learning models at large data limits. AtomSets also transfer better in a simulated materials discovery process where the targeted materials have property values out of the training data limits, require minimal domain knowledge inputs and are free from feature engineering. Check out this work here.